Example dataset
Sử dụng ví dụ ở chapter 10 trong cuốn sách này.
https://studyr.netlify.app/ref/introductory_biostatistics_2016.pdf
cancer <- read.csv("prostatecancer.dat", sep = "&")
cancer
Xray Grade Stage Age Acid Nodes
1 0 1 1 64 40 0
2 0 0 1 63 40 0
3 1 0 0 65 46 0
4 0 1 0 67 47 0
5 0 0 0 66 48 0
6 0 1 1 65 48 0
7 0 0 0 60 49 0
8 0 0 0 51 49 0
9 0 0 0 66 50 0
10 0 0 0 58 50 0
11 0 1 0 56 50 0
12 0 0 1 61 50 0
13 0 1 1 64 50 0
14 0 0 0 56 52 0
15 0 0 0 67 52 0
16 1 0 0 49 55 0
17 0 1 1 52 55 0
18 0 0 0 68 56 0
19 0 1 1 66 59 0
20 1 0 0 60 62 0
21 0 0 0 61 62 0
22 1 1 1 59 63 0
23 0 0 0 51 65 0
24 0 1 1 53 66 0
25 0 0 0 58 71 0
26 0 0 0 63 75 0
27 0 0 1 53 76 0
28 0 0 0 60 78 0
29 0 0 0 52 83 0
30 0 0 1 67 95 0
31 0 0 0 56 98 0
32 0 0 1 61 102 0
33 0 0 0 64 187 0
34 1 0 1 58 48 1
35 0 0 1 65 49 1
36 1 1 1 57 51 1
37 0 1 0 50 56 1
38 1 1 0 67 67 1
39 0 0 1 67 67 1
40 0 1 1 57 67 1
41 0 1 1 45 70 1
42 0 0 1 46 70 1
43 1 0 1 51 72 1
44 1 1 1 60 76 1
45 1 1 1 56 78 1
46 1 1 1 50 81 1
47 0 0 0 56 82 1
48 0 0 1 63 82 1
49 1 1 1 65 84 1
50 1 0 1 64 89 1
51 0 1 0 59 99 1
52 1 1 1 68 126 1
53 1 0 0 61 136 1
Logistic Regression Model
Effect of Measurement Scale
acid.glm <- glm(Nodes ~ Acid,
data = cancer,
family = binomial(link = "logit"))
# acid.glm
summary(acid.glm)
Call:
glm(formula = Nodes ~ Acid, family = binomial(link = "logit"),
data = cancer)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.92703 0.92104 -2.092 0.0364 *
Acid 0.02040 0.01257 1.624 0.1045
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 70.252 on 52 degrees of freedom
Residual deviance: 67.116 on 51 degrees of freedom
AIC: 71.116
Number of Fisher Scoring iterations: 4
or.50acid <- exp((100-50)*acid.glm$coefficients[2])
or.50acid
Testing Hypotheses in Multiple Logistic Regression
all.glm <- glm(Nodes ~ Xray + Grade + Stage + Age + Acid,
data = cancer,
family = binomial(link = "logit"))
summary(all.glm)
Call:
glm(formula = Nodes ~ Xray + Grade + Stage + Age + Acid, family = binomial(link = "logit"),
data = cancer)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.06180 3.45992 0.018 0.9857
Xray 2.04534 0.80718 2.534 0.0113 *
Grade 0.76142 0.77077 0.988 0.3232
Stage 1.56410 0.77401 2.021 0.0433 *
Age -0.06926 0.05788 -1.197 0.2314
Acid 0.02434 0.01316 1.850 0.0643 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 70.252 on 52 degrees of freedom
Residual deviance: 48.126 on 47 degrees of freedom
AIC: 60.126
Number of Fisher Scoring iterations: 5
chisq.LR <- all.glm$null.deviance - all.glm$deviance
chisq.LR
df.LR <- all.glm$df.null - all.glm$df.residual
df.LR
Stepwise
library(MASS)
MASS::stepAIC(all.glm)
Start: AIC=60.13
Nodes ~ Xray + Grade + Stage + Age + Acid
Df Deviance AIC
- Grade 1 49.097 59.097
- Age 1 49.615 59.615
<none> 48.126 60.126
- Acid 1 51.572 61.572
- Stage 1 52.558 62.558
- Xray 1 55.350 65.350
Step: AIC=59.1
Nodes ~ Xray + Stage + Age + Acid
Df Deviance AIC
- Age 1 50.660 58.660
<none> 49.097 59.097
- Acid 1 52.085 60.085
- Stage 1 55.381 63.381
- Xray 1 57.016 65.016
Step: AIC=58.66
Nodes ~ Xray + Stage + Acid
Df Deviance AIC
<none> 50.660 58.660
- Acid 1 53.353 59.353
- Stage 1 57.059 63.059
- Xray 1 58.613 64.613
Call: glm(formula = Nodes ~ Xray + Stage + Acid, family = binomial(link = "logit"),
data = cancer)
Coefficients:
(Intercept) Xray Stage Acid
-3.57565 2.06179 1.75556 0.02063
Degrees of Freedom: 52 Total (i.e. Null); 49 Residual
Null Deviance: 70.25
Residual Deviance: 50.66 AIC: 58.66
ROC curve
Loading required package: foreign
Loading required package: survival
Warning: package 'survival' was built under R version 4.4.1
Attaching package: 'survival'
The following object is masked _by_ '.GlobalEnv':
cancer
Loading required package: nnet
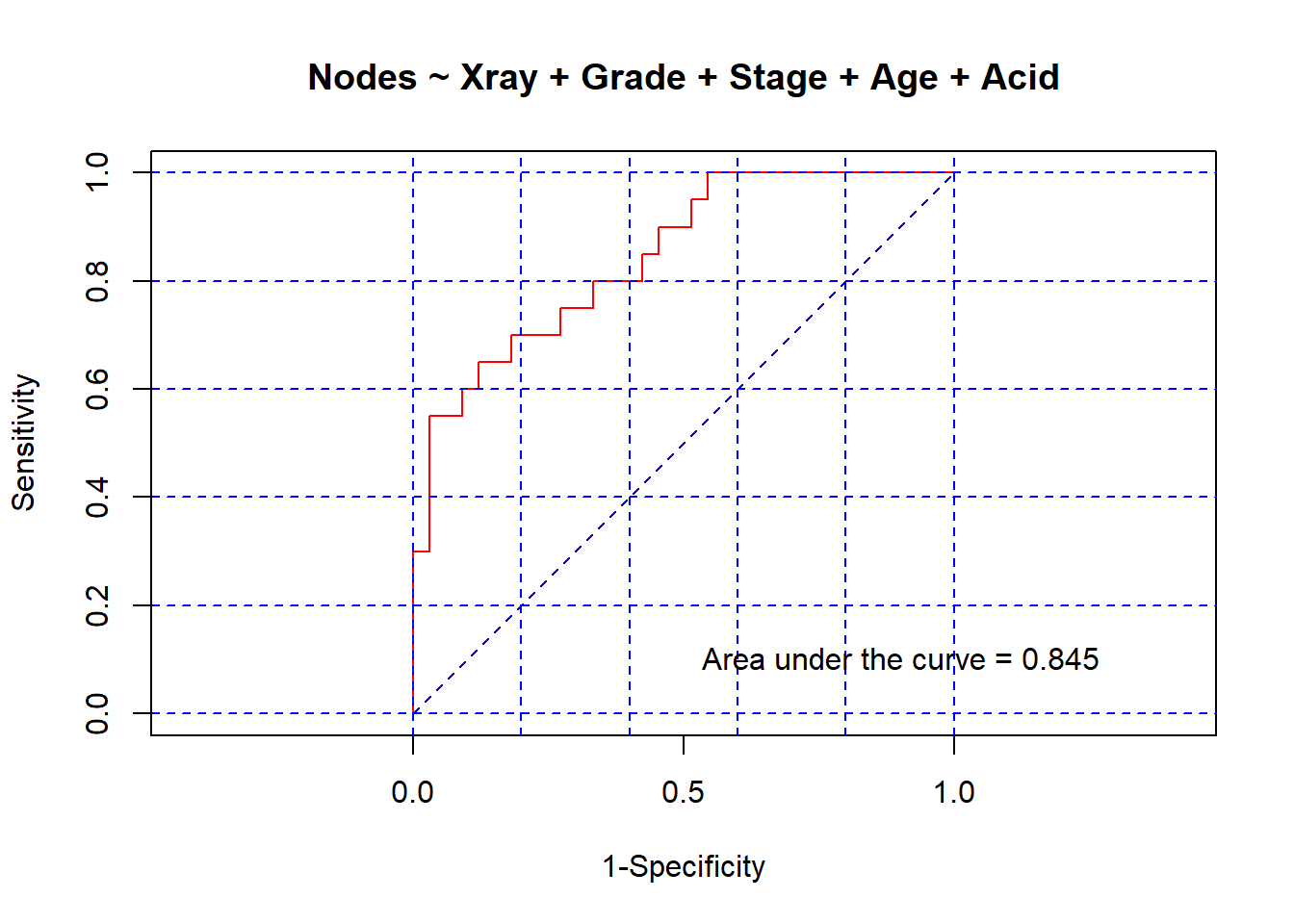
all.glm.roc <- lroc(all.glm, title = TRUE, auc.coords = c(0.5, 0.1))
$model.description
[1] "Nodes ~ Xray + Grade + Stage + Age + Acid"
$auc
[1] 0.8454545
$predicted.table
predicted.prob Non-diseased Diseased
0.0341 1 0
0.0351 1 0
0.0358 1 0
0.0361 1 0
0.0521 1 0
0.0607 1 0
0.0645 1 0
0.0657 1 0
0.0723 1 0
0.0775 1 0
0.0929 1 0
0.0973 1 0
0.1002 1 0
0.1314 1 0
0.1372 1 0
0.1393 0 1
0.1463 1 0
0.1566 0 1
0.1795 1 0
0.1929 1 0
0.2004 0 1
0.2007 1 0
0.2181 0 1
0.2184 1 0
0.2551 1 0
0.2796 1 0
0.2988 0 1
0.3040 1 0
0.3213 1 0
0.3227 0 1
0.3314 1 0
0.3684 1 0
0.4514 1 0
0.4648 0 1
0.4710 1 0
0.5130 1 0
0.5176 0 1
0.5311 1 0
0.5359 0 1
0.5452 1 0
0.5801 1 0
0.6948 0 1
0.7260 0 1
0.7673 0 1
0.8030 0 1
0.8489 0 1
0.8676 1 0
0.8689 0 1
0.8782 0 1
0.8935 0 1
0.9207 0 1
0.9421 0 1
0.9498 0 1
$diagnostic.table
1-Specificity Sensitivity
1.00000000 1.00
> 0.96969697 1.00
> 0.93939394 1.00
> 0.90909091 1.00
> 0.87878788 1.00
> 0.84848485 1.00
> 0.81818182 1.00
> 0.78787879 1.00
> 0.75757576 1.00
> 0.72727273 1.00
> 0.69696970 1.00
> 0.66666667 1.00
> 0.63636364 1.00
> 0.60606061 1.00
> 0.57575758 1.00
> 0.54545455 1.00
> 0.54545455 0.95
> 0.51515152 0.95
> 0.51515152 0.90
> 0.48484848 0.90
> 0.45454545 0.90
> 0.45454545 0.85
> 0.42424242 0.85
> 0.42424242 0.80
> 0.39393939 0.80
> 0.36363636 0.80
> 0.33333333 0.80
> 0.33333333 0.75
> 0.30303030 0.75
> 0.27272727 0.75
> 0.27272727 0.70
> 0.24242424 0.70
> 0.21212121 0.70
> 0.18181818 0.70
> 0.18181818 0.65
> 0.15151515 0.65
> 0.12121212 0.65
> 0.12121212 0.60
> 0.09090909 0.60
> 0.09090909 0.55
> 0.06060606 0.55
> 0.03030303 0.55
> 0.03030303 0.50
> 0.03030303 0.45
> 0.03030303 0.40
> 0.03030303 0.35
> 0.03030303 0.30
> 0.00000000 0.30
> 0.00000000 0.25
> 0.00000000 0.20
> 0.00000000 0.15
> 0.00000000 0.10
> 0.00000000 0.05
> 0.00000000 0.00